A stateless class for operations on cell data. Provides methods for: More...
#include <Intrepid2_CellTools.hpp>
Public Member Functions | |
| CellTools ()=default | |
| Default constructor. | |
| ~CellTools ()=default | |
| Destructor. | |
| template<typename InCellViewType , typename InputViewType> | |
| void | checkPointwiseInclusion (InCellViewType inCell, const InputViewType points, const shards::CellTopology cellTopo, const typename ScalarTraits< typename InputViewType::value_type >::scalar_type threshold) |
| template<typename Scalar > | |
| void | setJacobianDividedByDet (Data< Scalar, DeviceType > &jacobianDividedByDet, const Data< Scalar, DeviceType > &jacobian, const Data< Scalar, DeviceType > &jacobianDetInv) |
| template<typename PhysPointViewType , typename RefPointViewType , typename WorksetType , typename HGradBasisPtrType > | |
| void | mapToPhysicalFrame (PhysPointViewType physPoints, const RefPointViewType refPoints, const WorksetType worksetCell, const HGradBasisPtrType basis) |
| template<typename refSubcellViewType , typename paramViewType > | |
| void | mapToReferenceSubcell (refSubcellViewType refSubcellPoints, const paramViewType paramPoints, const ordinal_type subcellDim, const ordinal_type subcellOrd, const shards::CellTopology parentCell) |
| template<typename refSubcellViewType , typename paramViewType > | |
| void | mapToReferenceSubcell (refSubcellViewType refSubcellPoints, const paramViewType paramPoints, const typename RefSubcellParametrization< DeviceType >::ConstViewType subcellParametrization, const ordinal_type subcellOrd) |
| template<typename refSubcellViewType , typename paramViewType , typename ordViewType > | |
| void | mapToReferenceSubcell (refSubcellViewType refSubcellPoints, const paramViewType paramPoints, const typename RefSubcellParametrization< DeviceType >::ConstViewType subcellParametrization, const ordViewType subcellOrd) |
Static Public Member Functions | |
| static bool | hasReferenceCell (const shards::CellTopology cellTopo) |
| Checks if a cell topology has reference cell. More... | |
| template<typename JacobianViewType , typename PointViewType , typename WorksetType , typename HGradBasisType > | |
| static void | setJacobian (JacobianViewType jacobian, const PointViewType points, const WorksetType worksetCell, const Teuchos::RCP< HGradBasisType > basis, const int startCell=0, const int endCell=-1) |
| Computes the Jacobian matrix DF of the reference-to-physical frame map F. More... | |
| template<typename JacobianViewType , typename BasisGradientsType , typename WorksetType > | |
| static void | setJacobian (JacobianViewType jacobian, const WorksetType worksetCell, const BasisGradientsType gradients, const int startCell=0, const int endCell=-1) |
| Computes the Jacobian matrix DF of the reference-to-physical frame map F. More... | |
| template<typename JacobianViewType , typename PointViewType , typename WorksetCellViewType > | |
| static void | setJacobian (JacobianViewType jacobian, const PointViewType points, const WorksetCellViewType worksetCell, const shards::CellTopology cellTopo) |
| Computes the Jacobian matrix DF of the reference-to-physical frame map F. More... | |
| template<typename JacobianInvViewType , typename JacobianViewType > | |
| static void | setJacobianInv (JacobianInvViewType jacobianInv, const JacobianViewType jacobian) |
| Computes the inverse of the Jacobian matrix DF of the reference-to-physical frame map F. More... | |
| template<typename JacobianDetViewType , typename JacobianViewType > | |
| static void | setJacobianDet (JacobianDetViewType jacobianDet, const JacobianViewType jacobian) |
| Computes the determinant of the Jacobian matrix DF of the reference-to-physical frame map F. More... | |
| template<class PointScalar > | |
| static Data< PointScalar, DeviceType > | allocateJacobianDet (const Data< PointScalar, DeviceType > &jacobian) |
| Allocates and returns a Data container suitable for storing determinants corresponding to the Jacobians in the Data container provided. More... | |
| template<class PointScalar > | |
| static Data< PointScalar, DeviceType > | allocateJacobianInv (const Data< PointScalar, DeviceType > &jacobian) |
| Allocates and returns a Data container suitable for storing inverses corresponding to the Jacobians in the Data container provided. More... | |
| template<class PointScalar > | |
| static void | setJacobianDet (Data< PointScalar, DeviceType > &jacobianDet, const Data< PointScalar, DeviceType > &jacobian) |
| Computes determinants corresponding to the Jacobians in the Data container provided. More... | |
| template<class PointScalar > | |
| static void | setJacobianDetInv (Data< PointScalar, DeviceType > &jacobianDetInv, const Data< PointScalar, DeviceType > &jacobian) |
| Computes reciprocals of determinants corresponding to the Jacobians in the Data container provided. More... | |
| template<class PointScalar > | |
| static void | setJacobianInv (Data< PointScalar, DeviceType > &jacobianInv, const Data< PointScalar, DeviceType > &jacobian) |
| Computes determinants corresponding to the Jacobians in the Data container provided. More... | |
| template<class PointScalar > | |
| static void | setJacobianDividedByDet (Data< PointScalar, DeviceType > &jacobianDividedByDet, const Data< PointScalar, DeviceType > &jacobian, const Data< PointScalar, DeviceType > &jacobianDetInv) |
| Multiplies the Jacobian with shape (C,P,D,D) by the reciprocals of the determinants, with shape (C,P), entrywise. More... | |
| template<typename cellCenterValueType , class... cellCenterProperties> | |
| static void | getReferenceCellCenter (Kokkos::DynRankView< cellCenterValueType, cellCenterProperties...> cellCenter, const shards::CellTopology cell) |
| Computes the Cartesian coordinates of reference cell barycenter. More... | |
| template<typename cellVertexValueType , class... cellVertexProperties> | |
| static void | getReferenceVertex (Kokkos::DynRankView< cellVertexValueType, cellVertexProperties...> cellVertex, const shards::CellTopology cell, const ordinal_type vertexOrd) |
| Retrieves the Cartesian coordinates of a reference cell vertex. More... | |
| template<typename subcellVertexValueType , class... subcellVertexProperties> | |
| static void | getReferenceSubcellVertices (Kokkos::DynRankView< subcellVertexValueType, subcellVertexProperties...> subcellVertices, const ordinal_type subcellDim, const ordinal_type subcellOrd, const shards::CellTopology parentCell) |
| Retrieves the Cartesian coordinates of all vertices of a reference subcell. More... | |
| template<typename cellNodeValueType , class... cellNodeProperties> | |
| static void | getReferenceNode (Kokkos::DynRankView< cellNodeValueType, cellNodeProperties...> cellNode, const shards::CellTopology cell, const ordinal_type nodeOrd) |
| Retrieves the Cartesian coordinates of a reference cell node. More... | |
| template<typename SubcellNodeViewType > | |
| static void | getReferenceSubcellNodes (SubcellNodeViewType subcellNodes, const ordinal_type subcellDim, const ordinal_type subcellOrd, const shards::CellTopology parentCell) |
| Retrieves the Cartesian coordinates of all nodes of a reference subcell. More... | |
| template<typename RefEdgeTangentViewType > | |
| static void | getReferenceEdgeTangent (RefEdgeTangentViewType refEdgeTangent, const ordinal_type edgeOrd, const shards::CellTopology parentCell) |
| Computes constant tangent vectors to edges of 2D or 3D reference cells. More... | |
| template<typename RefFaceTanViewType > | |
| static void | getReferenceFaceTangents (RefFaceTanViewType refFaceTanU, RefFaceTanViewType refFaceTanV, const ordinal_type faceOrd, const shards::CellTopology parentCell) |
| Computes pairs of constant tangent vectors to faces of a 3D reference cells. More... | |
| template<typename RefSideNormalViewType > | |
| static void | getReferenceSideNormal (RefSideNormalViewType refSideNormal, const ordinal_type sideOrd, const shards::CellTopology parentCell) |
| Computes constant normal vectors to sides of 2D or 3D reference cells. More... | |
| template<typename RefFaceNormalViewType > | |
| static void | getReferenceFaceNormal (RefFaceNormalViewType refFaceNormal, const ordinal_type faceOrd, const shards::CellTopology parentCell) |
| Computes constant normal vectors to faces of 3D reference cell. More... | |
| template<typename edgeTangentValueType , class... edgeTangentProperties, typename worksetJacobianValueType , class... worksetJacobianProperties> | |
| static void | getPhysicalEdgeTangents (Kokkos::DynRankView< edgeTangentValueType, edgeTangentProperties...> edgeTangents, const Kokkos::DynRankView< worksetJacobianValueType, worksetJacobianProperties...> worksetJacobians, const ordinal_type worksetEdgeOrd, const shards::CellTopology parentCell) |
Computes non-normalized tangent vectors to physical edges in an edge workset 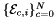 ; (see Subcell worksets for definition of edge worksets). More... ; (see Subcell worksets for definition of edge worksets). More... | |
| template<typename edgeTangentValueType , class... edgeTangentProperties, typename worksetJacobianValueType , class... worksetJacobianProperties, typename edgeOrdValueType , class... edgeOrdProperties> | |
| static void | getPhysicalEdgeTangents (Kokkos::DynRankView< edgeTangentValueType, edgeTangentProperties...> edgeTangents, const Kokkos::DynRankView< worksetJacobianValueType, worksetJacobianProperties...> worksetJacobians, const Kokkos::DynRankView< edgeOrdValueType, edgeOrdProperties...> worksetEdgeOrds, const shards::CellTopology parentCell) |
Computes non-normalized tangent vectors to physical edges in an edge workset 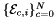 ; (see Subcell worksets for definition of edge worksets). More... ; (see Subcell worksets for definition of edge worksets). More... | |
| template<typename faceTanValueType , class... faceTanProperties, typename worksetJacobianValueType , class... worksetJacobianProperties> | |
| static void | getPhysicalFaceTangents (Kokkos::DynRankView< faceTanValueType, faceTanProperties...> faceTanU, Kokkos::DynRankView< faceTanValueType, faceTanProperties...> faceTanV, const Kokkos::DynRankView< worksetJacobianValueType, worksetJacobianProperties...> worksetJacobians, const ordinal_type worksetFaceOrd, const shards::CellTopology parentCell) |
Computes non-normalized tangent vector pairs to physical faces in a face workset 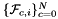 ; (see Subcell worksets for definition of face worksets). More... ; (see Subcell worksets for definition of face worksets). More... | |
| template<typename faceTanValueType , class... faceTanProperties, typename worksetJacobianValueType , class... worksetJacobianProperties, typename faceOrdValueType , class... faceOrdProperties> | |
| static void | getPhysicalFaceTangents (Kokkos::DynRankView< faceTanValueType, faceTanProperties...> faceTanU, Kokkos::DynRankView< faceTanValueType, faceTanProperties...> faceTanV, const Kokkos::DynRankView< worksetJacobianValueType, worksetJacobianProperties...> worksetJacobians, const Kokkos::DynRankView< faceOrdValueType, faceOrdProperties...> worksetFaceOrds, const shards::CellTopology parentCell) |
Computes non-normalized tangent vector pairs to physical faces in a face workset 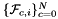 ; (see Subcell worksets for definition of face worksets). More... ; (see Subcell worksets for definition of face worksets). More... | |
| template<typename sideNormalValueType , class... sideNormalProperties, typename worksetJacobianValueType , class... worksetJacobianProperties> | |
| static void | getPhysicalSideNormals (Kokkos::DynRankView< sideNormalValueType, sideNormalProperties...> sideNormals, const Kokkos::DynRankView< worksetJacobianValueType, worksetJacobianProperties...> worksetJacobians, const ordinal_type worksetSideOrd, const shards::CellTopology parentCell) |
Computes non-normalized normal vectors to physical sides in a side workset 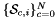 . More... . More... | |
| template<typename sideNormalValueType , class... sideNormalProperties, typename worksetJacobianValueType , class... worksetJacobianProperties, typename edgeOrdValueType , class... edgeOrdProperties> | |
| static void | getPhysicalSideNormals (Kokkos::DynRankView< sideNormalValueType, sideNormalProperties...> sideNormals, const Kokkos::DynRankView< worksetJacobianValueType, worksetJacobianProperties...> worksetJacobians, const Kokkos::DynRankView< edgeOrdValueType, edgeOrdProperties...> worksetSideOrds, const shards::CellTopology parentCell) |
Computes non-normalized normal vectors to physical sides in a side workset 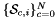 . It is similar to the CellTools::getPhysicalSideNormals function above, with the difference that the side ordinal can change from point to point, and it is provided by the rank-2 input array worksetSideOrds, indexed by (C,P). More... . It is similar to the CellTools::getPhysicalSideNormals function above, with the difference that the side ordinal can change from point to point, and it is provided by the rank-2 input array worksetSideOrds, indexed by (C,P). More... | |
| template<typename faceNormalValueType , class... faceNormalProperties, typename worksetJacobianValueType , class... worksetJacobianProperties> | |
| static void | getPhysicalFaceNormals (Kokkos::DynRankView< faceNormalValueType, faceNormalProperties...> faceNormals, const Kokkos::DynRankView< worksetJacobianValueType, worksetJacobianProperties...> worksetJacobians, const ordinal_type worksetFaceOrd, const shards::CellTopology parentCell) |
Computes non-normalized normal vectors to physical faces in a face workset 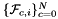 ; (see Subcell worksets for definition of face worksets). More... ; (see Subcell worksets for definition of face worksets). More... | |
| template<typename faceNormalValueType , class... faceNormalProperties, typename worksetJacobianValueType , class... worksetJacobianProperties, typename faceOrdValueType , class... faceOrdProperties> | |
| static void | getPhysicalFaceNormals (Kokkos::DynRankView< faceNormalValueType, faceNormalProperties...> faceNormals, const Kokkos::DynRankView< worksetJacobianValueType, worksetJacobianProperties...> worksetJacobians, const Kokkos::DynRankView< faceOrdValueType, faceOrdProperties...> worksetFaceOrds, const shards::CellTopology parentCell) |
Computes non-normalized normal vectors to physical faces in a face workset 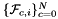 ; (see Subcell worksets for definition of face worksets). It is similar to the CellTools::getPhysicalSideNormals function above, with the difference that the side ordinal can change from point to point, and it is provided by the rank-2 input array worksetSideOrds, indexed by (C,P). More... ; (see Subcell worksets for definition of face worksets). It is similar to the CellTools::getPhysicalSideNormals function above, with the difference that the side ordinal can change from point to point, and it is provided by the rank-2 input array worksetSideOrds, indexed by (C,P). More... | |
| template<typename PhysPointValueType , typename RefPointValueType , typename WorksetType , typename HGradBasisPtrType > | |
| static void | mapToPhysicalFrame (PhysPointValueType physPoints, const RefPointValueType refPoints, const WorksetType worksetCell, const HGradBasisPtrType basis) |
| Computes F, the reference-to-physical frame map. More... | |
| template<typename PhysPointViewType , typename RefPointViewType , typename WorksetCellViewType > | |
| static void | mapToPhysicalFrame (PhysPointViewType physPoints, const RefPointViewType refPoints, const WorksetCellViewType worksetCell, const shards::CellTopology cellTopo) |
| Computes F, the reference-to-physical frame map. More... | |
| template<typename refSubcellViewType , typename paramPointViewType > | |
| static void | mapToReferenceSubcell (refSubcellViewType refSubcellPoints, const paramPointViewType paramPoints, const ordinal_type subcellDim, const ordinal_type subcellOrd, const shards::CellTopology parentCell) |
| Computes parameterization maps of 1- and 2-subcells of reference cells. More... | |
| template<typename refSubcellViewType , typename paramPointViewType > | |
| static void | mapToReferenceSubcell (refSubcellViewType refSubcellPoints, const paramPointViewType paramPoints, const typename RefSubcellParametrization< DeviceType >::ConstViewType subcellParametrization, const ordinal_type subcellOrd) |
| Computes parameterization maps of 1- and 2-subcells of reference cells. More... | |
| template<typename refSubcellViewType , typename paramPointViewType , typename ordViewType > | |
| static void | mapToReferenceSubcell (refSubcellViewType refSubcellPoints, const paramPointViewType paramPoints, const typename RefSubcellParametrization< DeviceType >::ConstViewType subcellParametrization, const ordViewType subcellOrd) |
| Computes parameterization maps of 1- and 2-subcells of reference cells. More... | |
| template<typename refPointValueType , class... refPointProperties, typename physPointValueType , class... physPointProperties, typename worksetCellValueType , class... worksetCellProperties> | |
| static void | mapToReferenceFrame (Kokkos::DynRankView< refPointValueType, refPointProperties...> refPoints, const Kokkos::DynRankView< physPointValueType, physPointProperties...> physPoints, const Kokkos::DynRankView< worksetCellValueType, worksetCellProperties...> worksetCell, const shards::CellTopology cellTopo) |
Computes 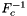 , the inverse of the reference-to-physical frame map using a default initial guess. More... , the inverse of the reference-to-physical frame map using a default initial guess. More... | |
| template<typename refPointValueType , class... refPointProperties, typename initGuessValueType , class... initGuessProperties, typename physPointValueType , class... physPointProperties, typename worksetCellValueType , class... worksetCellProperties, typename HGradBasisPtrType > | |
| static void | mapToReferenceFrameInitGuess (Kokkos::DynRankView< refPointValueType, refPointProperties...> refPoints, const Kokkos::DynRankView< initGuessValueType, initGuessProperties...> initGuess, const Kokkos::DynRankView< physPointValueType, physPointProperties...> physPoints, const Kokkos::DynRankView< worksetCellValueType, worksetCellProperties...> worksetCell, const HGradBasisPtrType basis) |
Computation of 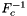 , the inverse of the reference-to-physical frame map using user-supplied initial guess. More... , the inverse of the reference-to-physical frame map using user-supplied initial guess. More... | |
| template<typename refPointValueType , class... refPointProperties, typename initGuessValueType , class... initGuessProperties, typename physPointValueType , class... physPointProperties, typename worksetCellValueType , class... worksetCellProperties> | |
| static void | mapToReferenceFrameInitGuess (Kokkos::DynRankView< refPointValueType, refPointProperties...> refPoints, const Kokkos::DynRankView< initGuessValueType, initGuessProperties...> initGuess, const Kokkos::DynRankView< physPointValueType, physPointProperties...> physPoints, const Kokkos::DynRankView< worksetCellValueType, worksetCellProperties...> worksetCell, const shards::CellTopology cellTopo) |
Computation of 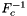 , the inverse of the reference-to-physical frame map using user-supplied initial guess. More... , the inverse of the reference-to-physical frame map using user-supplied initial guess. More... | |
| template<typename subcvCoordValueType , class... subcvCoordProperties, typename cellCoordValueType , class... cellCoordProperties> | |
| static void | getSubcvCoords (Kokkos::DynRankView< subcvCoordValueType, subcvCoordProperties...> subcvCoords, const Kokkos::DynRankView< cellCoordValueType, cellCoordProperties...> cellCoords, const shards::CellTopology primaryCell) |
| Computes coordinates of sub-control volumes in each primary cell. More... | |
| template<typename PointViewType > | |
| static bool | checkPointInclusion (const PointViewType point, const shards::CellTopology cellTopo, const typename ScalarTraits< typename PointViewType::value_type >::scalar_type thres=threshold< typename ScalarTraits< typename PointViewType::value_type >::scalar_type >()) |
| Checks if a point belongs to a reference cell. More... | |
| template<unsigned cellTopologyKey, typename OutputViewType , typename InputViewType > | |
| static void | checkPointwiseInclusion (OutputViewType inCell, const InputViewType points, const typename ScalarTraits< typename InputViewType::value_type >::scalar_type thresh=threshold< typename ScalarTraits< typename InputViewType::value_type >::scalar_type >()) |
| Checks every point for inclusion in the reference cell of a given topology. The points can belong to a global set and stored in a rank-2 view (P,D) , or to multiple sets indexed by a cell ordinal and stored in a rank-3 view (C,P,D). The cell topology key is a template argument. Requires cell topology with a reference cell. More... | |
| template<typename InCellViewType , typename PointViewType > | |
| static void | checkPointwiseInclusion (InCellViewType inCell, const PointViewType points, const shards::CellTopology cellTopo, const typename ScalarTraits< typename PointViewType::value_type >::scalar_type thres=threshold< typename ScalarTraits< typename PointViewType::value_type >::scalar_type >()) |
| Checks every point in multiple sets indexed by a cell ordinal for inclusion in the reference cell of a given topology. Requires cell topology with a reference cell. More... | |
| template<typename inCellValueType , class... inCellProperties, typename pointValueType , class... pointProperties, typename cellWorksetValueType , class... cellWorksetProperties> | |
| static void | checkPointwiseInclusion (Kokkos::DynRankView< inCellValueType, inCellProperties...> inCell, const Kokkos::DynRankView< pointValueType, pointProperties...> points, const Kokkos::DynRankView< cellWorksetValueType, cellWorksetProperties...> cellWorkset, const shards::CellTopology cellTopo, const typename ScalarTraits< pointValueType >::scalar_type thres=threshold< typename ScalarTraits< pointValueType >::scalar_type >()) |
| Checks every points for inclusion in physical cells from a cell workset. The points can belong to a global set and stored in a rank-2 (P,D) view, or to multiple sets indexed by a cell ordinal and stored in a rank-3 (C,P,D) view cells in a cell workset. More... | |
Private Types | |
| using | ExecSpaceType = typename DeviceType::execution_space |
| using | MemSpaceType = typename DeviceType::memory_space |
Static Private Member Functions | |
| template<typename outputValueType , typename pointValueType > | |
| static Teuchos::RCP< Basis < DeviceType, outputValueType, pointValueType > > | createHGradBasis (const shards::CellTopology cellTopo) |
| Generates default HGrad basis based on cell topology. More... | |
Detailed Description
template<typename DeviceType>
class Intrepid2::CellTools< DeviceType >
A stateless class for operations on cell data. Provides methods for:
- computing Jacobians of reference-to-physical frame mappings, their inverses and determinants
- application of the reference-to-physical frame mapping and its inverse
- parametrizations of edges and faces of reference cells needed for edge and face integrals,
- computation of edge and face tangents and face normals on both reference and physical frames
- inclusion tests for point sets in reference and physical cells.
Definition at line 77 of file Intrepid2_CellTools.hpp.
Member Function Documentation
|
static |
Allocates and returns a Data container suitable for storing determinants corresponding to the Jacobians in the Data container provided.
- Parameters
-
jacobian [in] - data with shape (C,P,D,D), as returned by CellGeometry::allocateJacobianData()
- Returns
- Data container with shape (C,P)
Definition at line 92 of file Intrepid2_CellToolsDefJacobian.hpp.
References Intrepid2::Data< DataScalar, DeviceType >::getExtents(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView1(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView3(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView4(), and Intrepid2::Data< DataScalar, DeviceType >::getVariationTypes().
|
static |
Allocates and returns a Data container suitable for storing inverses corresponding to the Jacobians in the Data container provided.
- Parameters
-
jacobian [in] - data with shape (C,P,D,D), as returned by CellGeometry::allocateJacobianData()
- Returns
- Data container with shape (C,P,D,D)
Definition at line 126 of file Intrepid2_CellToolsDefJacobian.hpp.
References Intrepid2::Data< DataScalar, DeviceType >::getExtents(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView1(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView2(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView3(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView4(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingViewRank(), Intrepid2::Data< DataScalar, DeviceType >::getVariationTypes(), and INTREPID2_TEST_FOR_EXCEPTION_DEVICE_SAFE.
|
static |
Checks if a point belongs to a reference cell.
Requires cell topology with a reference cell.
- Parameters
-
point [in] - rank-1 view (D) of the point tested for inclusion cellTopo [in] - cell topology threshold [in] - "tightness" of the inclusion test
- Returns
- true if the point is in the closure of the specified reference cell and false otherwise.
Definition at line 37 of file Intrepid2_CellToolsDefInclusion.hpp.
|
static |
Checks every point for inclusion in the reference cell of a given topology. The points can belong to a global set and stored in a rank-2 view (P,D) , or to multiple sets indexed by a cell ordinal and stored in a rank-3 view (C,P,D). The cell topology key is a template argument. Requires cell topology with a reference cell.
- Parameters
-
inCell [out] - rank-1 view (P) or rank-2 view (C,P). On return, its entries will be set to 1 or 0 depending on whether points are included in cells point [in] - rank-2 view (P,D) or rank-3 view (C,P,D) with reference coordinates of the points tested for inclusion threshold [in] - "tightness" of the inclusion test
Definition at line 131 of file Intrepid2_CellToolsDefInclusion.hpp.
|
static |
Checks every point in multiple sets indexed by a cell ordinal for inclusion in the reference cell of a given topology. Requires cell topology with a reference cell.
- Parameters
-
inRefCell [out] - rank-2 view (C,P) with results from the pointwise inclusion test refPoints [in] - rank-3 view (C,P,D) cellTopo [in] - cell topology threshold [in] - "tightness" of the inclusion test
|
static |
Checks every points for inclusion in physical cells from a cell workset. The points can belong to a global set and stored in a rank-2 (P,D) view, or to multiple sets indexed by a cell ordinal and stored in a rank-3 (C,P,D) view cells in a cell workset.
- Parameters
-
inCell [out] - rank-2 view (P,D) with results from the pointwise inclusion test points [in] - rank-2 view (P,D) or rank-3 view (C,P,D) with the physical points cellWorkset [in] - rank-3 view with dimensions (C,N,D) with the nodes of the cell workset cellTopo [in] - cell topology threshold [in] - tolerance for inclusion tests on the input points
Definition at line 216 of file Intrepid2_CellToolsDefInclusion.hpp.
References Intrepid2::RealSpaceTools< DeviceType >::clone().
|
inlinestaticprivate |
Generates default HGrad basis based on cell topology.
- Parameters
-
cellTopo [in] - cell topology
Definition at line 100 of file Intrepid2_CellTools.hpp.
|
static |
Computes non-normalized tangent vectors to physical edges in an edge workset 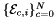 ; (see Subcell worksets for definition of edge worksets).
; (see Subcell worksets for definition of edge worksets).
For every edge in the workset the tangents are computed at the points 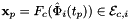 that are images of points from R=[-1,1] on edge
that are images of points from R=[-1,1] on edge 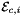 . Returns rank-3 array with dimensions (C,P,D1), D1=2 or D1=3 such that
. Returns rank-3 array with dimensions (C,P,D1), D1=2 or D1=3 such that
![\[ {edgeTangents}(c,p,d) = DF_c(\hat{\Phi}_i(t_p))\cdot {\partial{\hat{\Phi}}_{i}(t_p)\over\partial t}\,; \qquad t_p \in R \]](form_35.png)
In this formula:
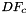 is Jacobian of parent cell
is Jacobian of parent cell 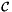 that owns physical edge
that owns physical edge 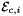 ;
; 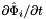 is the (constant) tangent to reference edge
is the (constant) tangent to reference edge 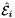 ; see Intrepid2::CellTools::getReferenceEdgeTangent that has the same local ordinal as the edges in the workset;
; see Intrepid2::CellTools::getReferenceEdgeTangent that has the same local ordinal as the edges in the workset; 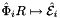 is parametrization of reference edge
is parametrization of reference edge 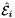 ;
;
- Warning
worksetJacobiansmust contain the values of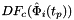 , where
, where ![$ t_p \in R=[-1,1] $](form_42.png) , i.e., Jacobians of the parent cells evaluated at points that are located on reference edge
, i.e., Jacobians of the parent cells evaluated at points that are located on reference edge 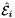 having the same local ordinal as the edges in the workset.
having the same local ordinal as the edges in the workset.
- Parameters
-
edgeTangents [out] - rank-3 array (C,P,D1) with tangents on workset edges worksetJacobians [in] - rank-4 array (C,P,D1,D1) with Jacobians evaluated at ref. edge points worksetEdgeOrd [in] - edge ordinal, relative to ref. cell, of the edge workset parentCell [in] - cell topology of the parent reference cell
Definition at line 372 of file Intrepid2_CellToolsDefNodeInfo.hpp.
References Intrepid2::RealSpaceTools< DeviceType >::matvec().
Referenced by Intrepid2::FunctionSpaceTools< DeviceType >::computeEdgeMeasure().
|
static |
Computes non-normalized tangent vectors to physical edges in an edge workset 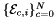 ; (see Subcell worksets for definition of edge worksets).
; (see Subcell worksets for definition of edge worksets).
It is similar to the CellTools::getPhysicalEdgeTangents function above, with the difference that the edge ordinal can change from point to point, and it is provided by the rank-2 input array worksetEdgeOrds, indexed by (C,P).
- Parameters
-
edgeTangents [out] - rank-3 array (C,P,D1) with tangents on workset edges worksetJacobians [in] - rank-4 array (C,P,D1,D1) with Jacobians evaluated at ref. edge points worksetEdgeOrds [in] - rank-2 array (C,P) with edge ordinals, relative to ref. cell, of the edge workset parentCell [in] - cell topology of the parent reference cell
Definition at line 454 of file Intrepid2_CellToolsDefNodeInfo.hpp.
References Intrepid2::RefSubcellParametrization< DeviceType >::get(), and Intrepid2::RealSpaceTools< DeviceType >::matvec().
|
static |
Computes non-normalized normal vectors to physical faces in a face workset 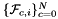 ; (see Subcell worksets for definition of face worksets).
; (see Subcell worksets for definition of face worksets).
For every face in the workset the normals are computed at the points 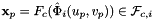 that are images of points from the parametrization domain R on face
that are images of points from the parametrization domain R on face 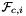 . Returns rank-3 array with dimensions (C,P,D), D=3, such that
. Returns rank-3 array with dimensions (C,P,D), D=3, such that
![\[ {faceNormals}(c,p,d) = \left( DF_c(\hat{\Phi}_i(u_p, v_p))\cdot {\partial\hat{\Phi}_i\over\partial u}\right) \times \left( DF_c(\hat{\Phi}_i(u_p, v_p))\cdot {\partial\hat{\Phi}_i\over\partial v}\right) \,; \qquad (u_p, v_p) \in R \,. \]](form_65.png)
In this formula:
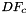 is Jacobian of parent cell
is Jacobian of parent cell 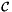 that owns physical face
that owns physical face 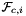 ;
; 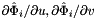 are the (constant) tangents on reference face
are the (constant) tangents on reference face 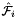 ; see Intrepid2::CellTools::getReferenceFaceTangents; that has the same local ordinal as the faces in the workset;
; see Intrepid2::CellTools::getReferenceFaceTangents; that has the same local ordinal as the faces in the workset; 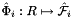 is parametrization of reference face
is parametrization of reference face 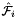 ;
; - R is the parametrization domain for reference face
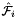 :
: ![\[ R = \left\{\begin{array}{rl} \{(0,0),(1,0),(0,1)\} & \mbox{if $\hat{\mathcal F}_i$ is Triangle} \\[1ex] [-1,1]\times [-1,1] & \mbox{if $\hat{\mathcal F}_i$ is Quadrilateral} \end{array}\right. \]](form_50.png)
- Warning
worksetJacobiansmust contain the values of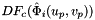 , where
, where 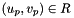 , i.e., Jacobians of the parent cells evaluated at points that are located on reference face
, i.e., Jacobians of the parent cells evaluated at points that are located on reference face 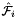 having the same local ordinal as the faces in the workset.
having the same local ordinal as the faces in the workset.
- Parameters
-
faceNormals [out] - rank-3 array (C,P,D), normals at workset faces worksetJacobians [in] - rank-4 array (C,P,D,D) with Jacobians at ref. face points worksetFaceOrd [in] - face ordinal, relative to ref. cell, of the face workset parentCell [in] - cell topology of the parent reference cell
Definition at line 813 of file Intrepid2_CellToolsDefNodeInfo.hpp.
References Intrepid2::RealSpaceTools< DeviceType >::vecprod().
Referenced by Intrepid2::FunctionSpaceTools< DeviceType >::computeFaceMeasure().
|
static |
Computes non-normalized normal vectors to physical faces in a face workset 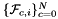 ; (see Subcell worksets for definition of face worksets). It is similar to the CellTools::getPhysicalSideNormals function above, with the difference that the side ordinal can change from point to point, and it is provided by the rank-2 input array worksetSideOrds, indexed by (C,P).
; (see Subcell worksets for definition of face worksets). It is similar to the CellTools::getPhysicalSideNormals function above, with the difference that the side ordinal can change from point to point, and it is provided by the rank-2 input array worksetSideOrds, indexed by (C,P).
- Parameters
-
faceNormals [out] - rank-3 array (C,P,D), normals at workset faces worksetJacobians [in] - rank-4 array (C,P,D,D) with Jacobians at ref. face points worksetFaceOrds [in] - rank-2 array (C,P) with face ordinals, relative to ref. cell, of the face workset parentCell [in] - cell topology of the parent reference cell
Definition at line 875 of file Intrepid2_CellToolsDefNodeInfo.hpp.
References Intrepid2::RealSpaceTools< DeviceType >::vecprod().
|
static |
Computes non-normalized tangent vector pairs to physical faces in a face workset 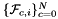 ; (see Subcell worksets for definition of face worksets).
; (see Subcell worksets for definition of face worksets).
For every face in the workset the tangents are computed at the points 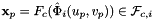 that are images of points from the parametrization domain R on face
that are images of points from the parametrization domain R on face 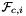 . Returns 2 rank-3 arrays with dimensions (C,P,D), D=3 such that
. Returns 2 rank-3 arrays with dimensions (C,P,D), D=3 such that
![\[ {faceTanU}(c,p,d) = DF_c(\hat{\Phi}_i(u_p, v_p))\cdot {\partial\hat{\Phi}_i\over\partial u};\qquad {faceTanV}(c,p,d) = DF_c(\hat{\Phi}_i(u_p, v_p))\cdot {\partial\hat{\Phi}_{i}\over\partial v}\,; \qquad (u_p, v_p) \in R \,. \]](form_46.png)
In this formula:
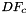 is Jacobian of parent cell
is Jacobian of parent cell 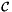 that owns physical face
that owns physical face 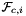 ;
; 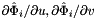 are the (constant) tangents on reference face
are the (constant) tangents on reference face 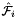 ; see Intrepid2::CellTools::getReferenceFaceTangents; that has the same local ordinal as the faces in the workset;
; see Intrepid2::CellTools::getReferenceFaceTangents; that has the same local ordinal as the faces in the workset; 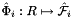 is parametrization of reference face
is parametrization of reference face 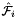 ;
; - R is the parametrization domain for reference face
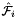 :
: ![\[ R = \left\{\begin{array}{rl} \{(0,0),(1,0),(0,1)\} & \mbox{if $\hat{\mathcal F}_i$ is Triangle} \\[1ex] [-1,1]\times [-1,1] & \mbox{if $\hat{\mathcal F}_i$ is Quadrilateral} \end{array}\right. \]](form_50.png)
- Warning
worksetJacobiansmust contain the values of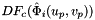 , where
, where 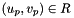 , i.e., Jacobians of the parent cells evaluated at points that are located on reference face
, i.e., Jacobians of the parent cells evaluated at points that are located on reference face 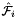 having the same local ordinal as the faces in the workset.
having the same local ordinal as the faces in the workset.
- Parameters
-
faceTanU [out] - rank-3 array (C,P,D), image of ref. face u-tangent at workset faces faceTanV [out] - rank-3 array (C,P,D), image of ref. face u-tangent at workset faces worksetJacobians [in] - rank-4 array (C,P,D,D) with Jacobians at ref. face points worksetFaceOrd [in] - face ordinal, relative to ref. cell, of the face workset parentCell [in] - cell topology of the parent reference cell
Definition at line 518 of file Intrepid2_CellToolsDefNodeInfo.hpp.
References Intrepid2::RealSpaceTools< DeviceType >::matvec().
|
static |
Computes non-normalized tangent vector pairs to physical faces in a face workset 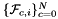 ; (see Subcell worksets for definition of face worksets).
; (see Subcell worksets for definition of face worksets).
It is similar to the CellTools::getPhysicalFaceTangents function above, with the difference that the face ordinal can change from point to point, and it is provided by the rank-2 input array worksetFaceOrds, indexed by (C,P).
- Parameters
-
faceTanU [out] - rank-3 array (C,P,D), image of ref. face u-tangent at workset faces faceTanV [out] - rank-3 array (C,P,D), image of ref. face u-tangent at workset faces worksetJacobians [in] - rank-4 array (C,P,D,D) with Jacobians at ref. face points worksetFaceOrds [in] - rank-2 array (C,P) with face ordinals, relative to ref. cell, of the face workset parentCell [in] - cell topology of the parent reference cell
Definition at line 614 of file Intrepid2_CellToolsDefNodeInfo.hpp.
References Intrepid2::RefSubcellParametrization< DeviceType >::get(), and Intrepid2::RealSpaceTools< DeviceType >::matvec().
|
static |
Computes non-normalized normal vectors to physical sides in a side workset 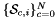 .
.
For every side in the workset the normals are computed at the points 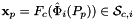 that are images of points from the parametrization domain R on side
that are images of points from the parametrization domain R on side 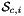 . A side is defined as a subcell of dimension one less than that of its parent cell. Therefore, sides of 2D cells are 1-subcells (edges) and sides of 3D cells are 2-subcells (faces).
. A side is defined as a subcell of dimension one less than that of its parent cell. Therefore, sides of 2D cells are 1-subcells (edges) and sides of 3D cells are 2-subcells (faces).
Returns rank-3 array with dimensions (C,P,D), D = 2 or 3, such that
![\[ {sideNormals}(c,p,d) = \left\{\begin{array}{crl} \displaystyle \left(DF_c(\hat{\Phi}_i(t_p))\cdot {\partial{\hat{\Phi}}_{i}(t_p)\over\partial t}\right)^{\perp} & t_p\in R & \mbox{for 2D parent cells} \\[2ex] \displaystyle \left( DF_c(\hat{\Phi}_i(u_p, v_p))\cdot {\partial\hat{\Phi}_i\over\partial u}\right) \times \left( DF_c(\hat{\Phi}_i(u_p, v_p))\cdot {\partial\hat{\Phi}_i\over\partial v}\right) \,; & (u_p, v_p) \in R & \mbox{for 3D parent cells} \end{array}\right. \]](form_56.png)
In this formula:
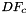 is Jacobian of parent cell
is Jacobian of parent cell 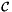 that owns physical side
that owns physical side 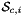 ;
; - For 2D cells:
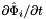 is the (constant) tangent to reference side (edge)
is the (constant) tangent to reference side (edge) 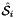 ; see Intrepid2::CellTools::getReferenceEdgeTangent, that has the same local ordinal as the sides in the workset;
; see Intrepid2::CellTools::getReferenceEdgeTangent, that has the same local ordinal as the sides in the workset; - For 3D cells:
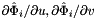 are the (constant) tangents on reference side (face)
are the (constant) tangents on reference side (face) 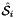 ; see Intrepid2::CellTools::getReferenceFaceTangents, that has the same local ordinal as the sides in the workset;
; see Intrepid2::CellTools::getReferenceFaceTangents, that has the same local ordinal as the sides in the workset; 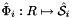 is parametrization of reference side
is parametrization of reference side 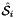 ;
; - R is the parametrization domain for reference side
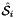 . For 2D parent cells R=[-1,1] and for 3D parent cells
. For 2D parent cells R=[-1,1] and for 3D parent cells ![\[ R = \left\{\begin{array}{rl} \{(0,0),(1,0),(0,1)\} & \mbox{if $\hat{\mathcal S}_i$ is Triangle} \\[1ex] [-1,1]\times [-1,1] & \mbox{if $\hat{\mathcal S}_i$ is Quadrilateral} \end{array}\right. \]](form_59.png)
- Remarks
- For 3D cells the physical side normals coincides with the face normals computed by Intrepid2::CellTools::getPhysicalFaceNormals and these two methods are completely equivalent.
- For 2D cells the physical side normals are defined by
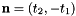 where
where 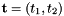 are the physical edge tangents computed by Intrepid2::CellTools::getPhysicalEdgeTangents. Therefore, the pairs
are the physical edge tangents computed by Intrepid2::CellTools::getPhysicalEdgeTangents. Therefore, the pairs 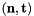 are positively oriented.
are positively oriented.
- Warning
worksetJacobiansmust contain the values of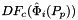 , where
, where 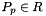 , i.e., Jacobians of the parent cells evaluated at points that are located on reference side
, i.e., Jacobians of the parent cells evaluated at points that are located on reference side 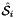 having the same local ordinal as the sides in the workset.
having the same local ordinal as the sides in the workset.
- Parameters
-
sideNormals [out] - rank-3 array (C,P,D), normals at workset sides worksetJacobians [in] - rank-4 array (C,P,D,D) with Jacobians at ref. side points worksetSideOrd [in] - side ordinal, relative to ref. cell, of the side workset parentCell [in] - cell topology of the parent reference cell
Definition at line 715 of file Intrepid2_CellToolsDefNodeInfo.hpp.
Referenced by Intrepid2::CubatureControlVolumeSide< DeviceType, pointValueType, weightValueType >::getCubature().
|
static |
Computes non-normalized normal vectors to physical sides in a side workset 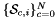 . It is similar to the CellTools::getPhysicalSideNormals function above, with the difference that the side ordinal can change from point to point, and it is provided by the rank-2 input array worksetSideOrds, indexed by (C,P).
. It is similar to the CellTools::getPhysicalSideNormals function above, with the difference that the side ordinal can change from point to point, and it is provided by the rank-2 input array worksetSideOrds, indexed by (C,P).
- Parameters
-
sideNormals [out] - rank-3 array (C,P,D), normals at workset sides worksetJacobians [in] - rank-4 array (C,P,D,D) with Jacobians at ref. side points worksetSideOrds [in] - rank-2 array (C,P) with side ordinals, relative to ref. cell, of the side workset parentCell [in] - cell topology of the parent reference cell
Definition at line 766 of file Intrepid2_CellToolsDefNodeInfo.hpp.
|
static |
Computes the Cartesian coordinates of reference cell barycenter.
Requires cell topology with a reference cell.
- Parameters
-
cellCenter [out] - coordinates of the specified reference cell center cell [in] - cell topology
Definition at line 36 of file Intrepid2_CellToolsDefNodeInfo.hpp.
References Intrepid2::RefCellCenter< DeviceType >::get().
Referenced by Intrepid2::Basis_HVOL_C0_FEM< DeviceType, outputValueType, pointValueType >::Basis_HVOL_C0_FEM().
|
static |
Computes constant tangent vectors to edges of 2D or 3D reference cells.
Returns rank-1 array with dimension (D), D=2 or D=3; such that
![\[ {refEdgeTangent}(*) = \hat{\bf t}_i = {\partial\hat{\Phi}_i(t)\over\partial t}\,, \]](form_5.png)
where ![$\hat{\Phi}_i : R =[-1,1]\mapsto \hat{\mathcal E}_i$](form_6.png) is the parametrization map of the specified reference edge
is the parametrization map of the specified reference edge 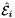 , given by
, given by
![\[ \hat{\Phi}_i(t) = \left\{\begin{array}{ll} (\hat{x}(t),\hat{y}(t),\hat{z}(t)) & \mbox{for 3D parent cells} \\[1ex] (\hat{x}(t),\hat{y}(t)) & \mbox{for 2D parent cells} \\[1ex] \end{array}\right. \]](form_8.png)
The length of computed edge tangents is one-half the length of their associated edges:
![\[ |\hat{\bf t}_i | = {1\over 2} |\hat{\mathcal E}_i |\,. \]](form_9.png)
Because the edges of all reference cells are always affine images of [-1,1], the edge tangent is constant vector field.
- Parameters
-
refEdgeTangent [out] - rank-1 array (D) with the edge tangent; D = cell dimension edgeOrd [in] - ordinal of the edge whose tangent is computed parentCell [in] - cell topology of the parent reference cell
Definition at line 211 of file Intrepid2_CellToolsDefNodeInfo.hpp.
References Intrepid2::RefSubcellParametrization< DeviceType >::get().
Referenced by Intrepid2::Basis_HCURL_TET_In_FEM< DeviceType, outputValueType, pointValueType >::Basis_HCURL_TET_In_FEM(), Intrepid2::Basis_HCURL_TRI_In_FEM< DeviceType, outputValueType, pointValueType >::Basis_HCURL_TRI_In_FEM(), Intrepid2::ProjectionTools< DeviceType >::getHCurlBasisCoeffs(), Intrepid2::ProjectionTools< DeviceType >::getHGradBasisCoeffs(), and Intrepid2::ProjectionTools< DeviceType >::getL2BasisCoeffs().
|
static |
Computes constant normal vectors to faces of 3D reference cell.
Returns rank-1 array with dimension (D), D=3 such that
![\[ {refFaceNormal}(*) = \hat{\bf n}_i = {\partial\hat{\Phi}_{i}\over\partial u} \times {\partial\hat{\Phi}_{i}\over\partial v} \]](form_28.png)
where 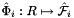 is the parametrization map of the specified reference face
is the parametrization map of the specified reference face 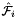 given by
given by
![\[ \hat{\Phi}_i(u,v) =(\hat{x}(u,v),\hat{y}(u,v),\hat{z}(u,v)) \]](form_14.png)
and
![\[ R = \left\{\begin{array}{rl} \{(0,0),(1,0),(0,1)\} & \mbox{if ${\mathcal F}$ is Triangle} \\[1ex] [-1,1]\times [-1,1] & \mbox{if ${\mathcal F}$ is Quadrilateral} \,. \end{array}\right. \]](form_29.png)
The length of computed face normals is proportional to face area:
![\[ |\hat{\bf n}_i | = \left\{\begin{array}{rl} 2 \mbox{Area}(\hat{\mathcal F}_i) & \mbox{if $\hat{\mathcal F}_i$ is Triangle} \\[1ex] \mbox{Area}(\hat{\mathcal F}_i) & \mbox{if $\hat{\mathcal F}_i$ is Quadrilateral} \,. \end{array}\right. \]](form_23.png)
Because the faces of all reference cells are always affine images of R , the coordinate functions 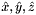 of the parametrization map are linear and the face normal is a constant vector.
of the parametrization map are linear and the face normal is a constant vector.
- Remarks
- The method Intrepid2::CellTools::getReferenceFaceTangents computes the reference face tangents
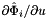 and
and 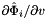 .
.
- Parameters
-
refFaceNormal [out] - rank-1 array (D) with (constant) face normal faceOrd [in] - ordinal of the face whose normal is computed parentCell [in] - cell topology of the parent reference cell
Definition at line 335 of file Intrepid2_CellToolsDefNodeInfo.hpp.
References Intrepid2::RealSpaceTools< DeviceType >::vecprod().
Referenced by Intrepid2::ProjectionTools< DeviceType >::getHCurlBasisCoeffs(), and Intrepid2::ProjectionTools< DeviceType >::getHGradBasisCoeffs().
|
static |
Computes pairs of constant tangent vectors to faces of a 3D reference cells.
Returns 2 rank-1 arrays with dimension (D), D=3, such that
![\[ {refFaceTanU}(*) = \hat{\bf t}_{i,u} = {\partial\hat{\Phi}_i(u,v)\over\partial u} = \left({\partial\hat{x}(u,v)\over\partial u}, {\partial\hat{y}(u,v)\over\partial u}, {\partial\hat{z}(u,v)\over\partial u} \right) ; \]](form_10.png)
![\[ {refFaceTanV}(*) = \hat{\bf t}_{i,v} = {\partial\hat{\Phi}_i(u,v)\over \partial v} = \left({\partial\hat{x}(u,v)\over\partial v}, {\partial\hat{y}(u,v)\over\partial v}, {\partial\hat{z}(u,v)\over\partial v} \right)\,; \]](form_11.png)
where 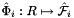 is the parametrization map of the specified reference face
is the parametrization map of the specified reference face 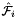 given by
given by
![\[ \hat{\Phi}_i(u,v) =(\hat{x}(u,v),\hat{y}(u,v),\hat{z}(u,v)) \]](form_14.png)
and
![\[ R = \left\{\begin{array}{rl} \{(0,0),(1,0),(0,1)\} & \mbox{if $\hat{\mathcal F}_i$ is Triangle} \\[1ex] [-1,1]\times [-1,1] & \mbox{if $\hat{\mathcal F}_i$ is Quadrilateral} \,. \end{array}\right. \]](form_15.png)
Because the faces of all reference cells are always affine images of R , the coordinate functions 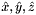 of the parametrization map are linear and the face tangents are constant vectors.
of the parametrization map are linear and the face tangents are constant vectors.
- Parameters
-
refFaceTanU [out] - rank-1 array (D) with (constant) tangent in u-direction refFaceTanV [out] - rank-1 array (D) with (constant) tangent in v-direction faceOrd [in] - ordinal of the face whose tangents are computed parentCell [in] - cell topology of the parent 3D reference cell
Definition at line 249 of file Intrepid2_CellToolsDefNodeInfo.hpp.
References Intrepid2::RefSubcellParametrization< DeviceType >::get().
Referenced by Intrepid2::Basis_HCURL_TET_In_FEM< DeviceType, outputValueType, pointValueType >::Basis_HCURL_TET_In_FEM().
|
static |
Retrieves the Cartesian coordinates of a reference cell node.
Returns Cartesian coordinates of a reference cell node. Requires cell topology with a reference cell. Node coordinates are always returned as an (x,y,z)-triple regardlesss of the actual topological cell dimension. The unused coordinates are set to zero, e.g., node 0 of Line<2> is returned as {-1,0,0}.
- Remarks
- Because the nodes of a cell with a base topology coincide with its vertices, for cells with base topology this method is equivalent to Intrepid2::CellTools::getReferenceVertex.
- Parameters
-
cellNode [out] - coordinates of the specified reference vertex cell [in] - cell topology of the cell vertexOrd [in] - node ordinal
Definition at line 139 of file Intrepid2_CellToolsDefNodeInfo.hpp.
References Intrepid2::RefCellNodes< DeviceType >::get().
|
static |
Computes constant normal vectors to sides of 2D or 3D reference cells.
A side is defined as a subcell of dimension one less than that of its parent cell. Therefore, sides of 2D cells are 1-subcells (edges) and sides of 3D cells are 2-subcells (faces).
Returns rank-1 array with dimension (D), D = 2 or 3 such that
![\[ {refSideNormal}(*) = \hat{\bf n}_i = \left\{\begin{array}{rl} \displaystyle \left({\partial\hat{\Phi}_i(t)\over\partial t}\right)^{\perp} & \mbox{for 2D parent cells} \\[2ex] \displaystyle {\partial\hat{\Phi}_{i}\over\partial u} \times {\partial\hat{\Phi}_{i}\over\partial v} & \mbox{for 3D parent cells} \end{array}\right. \]](form_17.png)
where  , and
, and 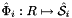 is the parametrization map of the specified reference side
is the parametrization map of the specified reference side 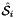 given by
given by
![\[ \hat{\Phi}_i(u,v) = \left\{\begin{array}{rl} (\hat{x}(t),\hat{y}(t)) & \mbox{for 2D parent cells} \\[1ex] (\hat{x}(u,v),\hat{y}(u,v),\hat{z}(u,v)) & \mbox{for 3D parent cells} \end{array}\right. \]](form_21.png)
For sides of 2D cells R=[-1,1] and for sides of 3D cells
![\[ R = \left\{\begin{array}{rl} \{(0,0),(1,0),(0,1)\} & \mbox{if $\hat{\mathcal S}_i$ is Triangle} \\[1ex] [-1,1]\times [-1,1] & \mbox{if $\hat{\mathcal S}_i$ is Quadrilateral} \,. \end{array}\right. \]](form_22.png)
For 3D cells the length of computed side normals is proportional to side area:
![\[ |\hat{\bf n}_i | = \left\{\begin{array}{rl} 2 \mbox{Area}(\hat{\mathcal F}_i) & \mbox{if $\hat{\mathcal F}_i$ is Triangle} \\[1ex] \mbox{Area}(\hat{\mathcal F}_i) & \mbox{if $\hat{\mathcal F}_i$ is Quadrilateral} \,. \end{array}\right. \]](form_23.png)
For 2D cells the length of computed side normals is proportional to side length:
![\[ |\hat{\bf n}_i | = {1\over 2} |\hat{\mathcal F}_i |\,. \]](form_24.png)
Because the sides of all reference cells are always affine images of R , the coordinate functions 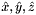 of the parametrization maps are linear and the side normal is a constant vector.
of the parametrization maps are linear and the side normal is a constant vector.
- Remarks
- For 3D cells the reference side normal coincides with the face normal computed by Intrepid2::CellTools::getReferenceFaceNormal and these two methods are completely equivalent.
- For 2D cells the reference side normal is defined by
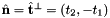 where
where 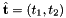 is the tangent vector computed by Intrepid2::CellTools::getReferenceEdgeTangent. Therefore, the pair
is the tangent vector computed by Intrepid2::CellTools::getReferenceEdgeTangent. Therefore, the pair 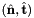 is positively oriented.
is positively oriented.
- Parameters
-
refSideNormal [out] - rank-1 array (D) with (constant) side normal sideOrd [in] - ordinal of the side whose normal is computed parentCell [in] - cell topology of the parent reference cell
Definition at line 294 of file Intrepid2_CellToolsDefNodeInfo.hpp.
Referenced by Intrepid2::Basis_HDIV_TET_In_FEM< DeviceType, outputValueType, pointValueType >::Basis_HDIV_TET_In_FEM(), Intrepid2::Basis_HDIV_TRI_In_FEM< DeviceType, outputValueType, pointValueType >::Basis_HDIV_TRI_In_FEM(), Intrepid2::ProjectionTools< DeviceType >::getHDivBasisCoeffs(), and Intrepid2::ProjectionTools< DeviceType >::getL2BasisCoeffs().
|
static |
Retrieves the Cartesian coordinates of all nodes of a reference subcell.
Returns rank-2 array with the Cartesian coordinates of the nodes of the specified reference cell subcell. Requires cell topology with a reference cell.
- Parameters
-
subcellNodes [out] - rank-2 (N,D) array with the Cartesian coordinates of the reference subcell nodes subcellDim [in] - dimension of the subcell; 0 <= subcellDim <= parentCell dimension subcellOrd [in] - ordinal of the subcell parentCell [in] - topology of the cell that owns the subcell
- Remarks
- When
subcellDim= dimension of theparentCellthis method returns the Cartesian coordinates of the nodes of the reference cell itself. Note that this requiressubcellOrd=0.
Definition at line 169 of file Intrepid2_CellToolsDefNodeInfo.hpp.
|
static |
Retrieves the Cartesian coordinates of all vertices of a reference subcell.
Returns rank-2 array with the Cartesian coordinates of the vertices of the specified reference cell subcell. Requires cell topology with a reference cell.
- Parameters
-
subcellVertices [out] - rank-2 (V,D) array with the Cartesian coordinates of the reference subcell vertices subcellDim [in] - dimension of the subcell; 0 <= subcellDim <= parentCell dimension subcellOrd [in] - ordinal of the subcell parentCell [in] - topology of the cell that owns the subcell
- Remarks
- When
subcellDim= dimension of theparentCellthis method returns the Cartesian coordinates of the vertices of the reference cell itself. Note that this requires subcellOrd=0.
Definition at line 103 of file Intrepid2_CellToolsDefNodeInfo.hpp.
|
static |
Retrieves the Cartesian coordinates of a reference cell vertex.
Requires cell topology with a reference cell.
- Parameters
-
cellVertex [out] - coordinates of the specified reference cell vertex cell [in] - cell topology of the cell vertexOrd [in] - vertex ordinal
Definition at line 68 of file Intrepid2_CellToolsDefNodeInfo.hpp.
References Intrepid2::RefCellNodes< DeviceType >::get().
Referenced by Intrepid2::ProjectionStruct< DeviceType, ValueType >::createHGradProjectionStruct(), and Intrepid2::ProjectionStruct< DeviceType, ValueType >::createL2ProjectionStruct().
|
static |
Computes coordinates of sub-control volumes in each primary cell.
To build the system of equations for the control volume finite element method we need to compute geometric data for integration over control volumes. A control volume is polygon or polyhedron that surrounds a primary cell node and has vertices that include the surrounding primary cells' barycenter, edge midpoints, and face midpoints if in 3-d.
When using element-based assembly of the discrete equations over the primary mesh, a single element will contain a piece of each control volume surrounding each of the primary cell nodes. This piece of control volume (sub-control volume) is always a quadrilateral in 2-d and a hexahedron in 3-d.
In 2-d the sub-control volumes are defined in the following way:
Quadrilateral primary element:
O________M________O
| | |
| 3 | 2 | B = cell barycenter
| | | O = primary cell nodes
M________B________M M = cell edge midpoints
| | |
| 0 | 1 | sub-control volumes 0, 1, 2, 3
| | |
O________M________O
Triangle primary element:
O
/ \
/ \ B = cell barycenter
/ \ O = primary cell nodes
M 2 M M = cell edge midpoints
/ \ / \
/ \ B / \ sub-control volumes 0, 1, 2
/ | \
/ 0 | 1 \
O________M________OIn 3-d the sub-control volumes are defined by the primary cell face centers and edge midpoints. The eight sub-control volumes for a hexahedron are shown below:
O__________E__________O
/| /| /|
E_|________F_|________E |
/| | /| | /| |
O_|_|______E_|_|______O | | O = primary cell nodes
| | E------|-|-F------|-|-E B = cell barycenter
| |/| | |/| | |/| F = cell face centers
| F-|------|-B-|------|-F | E = cell edge midpoints
|/| | |/| | |/| |
E_|_|______F_|_|______E | |
| | O------|-|-E------|-|-O
| |/ | |/ | |/
| E--------|-F--------|-E
|/ |/ |/
O__________E__________O- Parameters
-
subCVCoords [out] - array containing sub-control volume coordinates cellCoords [in] - array containing coordinates of primary cells primaryCell [in] - primary cell topology
Definition at line 340 of file Intrepid2_CellToolsDefControlVolume.hpp.
Referenced by Intrepid2::CubatureControlVolumeBoundary< DeviceType, pointValueType, weightValueType >::getCubature(), Intrepid2::CubatureControlVolume< DeviceType, pointValueType, weightValueType >::getCubature(), and Intrepid2::CubatureControlVolumeSide< DeviceType, pointValueType, weightValueType >::getCubature().
|
inlinestatic |
Checks if a cell topology has reference cell.
- Parameters
-
cell [in] - cell topology
- Returns
- true if the cell topology has reference cell, false otherwise
Definition at line 88 of file Intrepid2_CellTools.hpp.
References Intrepid2::RefSubcellParametrization< DeviceType >::isSupported().
|
static |
Computes F, the reference-to-physical frame map.
There are 2 use cases:
- Applies
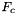 for all cells in a cell workset to a single point set stored in a rank-2 (P,D) array;
for all cells in a cell workset to a single point set stored in a rank-2 (P,D) array; - Applies
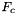 for all cells in a cell workset to multiple point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array;
for all cells in a cell workset to multiple point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array;
For a single point set in a rank-2 array (P,D) returns a rank-3 (C,P,D) array such that
![\[ \mbox{physPoints}(c,p,d) = \Big(F_c(\mbox{refPoint}(p,*)) \Big)_d \quad c=0,\ldots, C \]](form_67.png)
For multiple point sets in a rank-3 (C,P,D) array returns a rank-3 (C,P,D) array such that
![\[ \mbox{physPoints}(c,p,d) = \Big(F_c(\mbox{refPoint}(c,p,*)) \Big)_d \quad c=0,\ldots, C \]](form_68.png)
This corresponds to mapping multiple sets of reference points to a matching number of physical cells.
Requires pointer to HGrad basis that defines reference to physical cell mapping. See Section Reference-to-physical cell mapping for definition of the mapping function.
- Warning
- The array
refPointsrepresents an arbitrary set of points in the reference frame that are not required to be in the reference cell corresponding to the specified cell topology. As a result, the images of these points under a given reference-to-physical map are not necessarily contained in the physical cell that is the image of the reference cell under that map. CellTools provides several inclusion tests methods to check whether or not the points are inside a reference cell.
- Parameters
-
physPoints [out] - rank-3 array with dimensions (C,P,D) with the images of the ref. points refPoints [in] - rank-3/2 array with dimensions (C,P,D)/(P,D) with points in reference frame cellWorkset [in] - rank-3 container with logical dimensions (C,N,D) with the nodes of the cell workset basis [in] - pointer to HGrad basis used in reference-to-physical cell mapping
Referenced by Intrepid2::CellTools< DeviceType >::mapToPhysicalFrame(), and Intrepid2::ProjectedGeometry< spaceDim, PointScalar, DeviceType >::projectOntoHGRADBasis().
|
inlinestatic |
Computes F, the reference-to-physical frame map.
There are 2 use cases:
- Applies
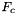 for all cells in a cell workset to a single point set stored in a rank-2 (P,D) array;
for all cells in a cell workset to a single point set stored in a rank-2 (P,D) array; - Applies
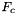 for all cells in a cell workset to multiple point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array;
for all cells in a cell workset to multiple point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array;
For a single point set in a rank-2 array (P,D) returns a rank-3 (C,P,D) array such that
![\[ \mbox{physPoints}(c,p,d) = \Big(F_c(\mbox{refPoint}(p,*)) \Big)_d \quad c=0,\ldots, C \]](form_67.png)
For multiple point sets in a rank-3 (C,P,D) array returns a rank-3 (C,P,D) array such that
![\[ \mbox{physPoints}(c,p,d) = \Big(F_c(\mbox{refPoint}(c,p,*)) \Big)_d \quad c=0,\ldots, C \]](form_68.png)
This corresponds to mapping multiple sets of reference points to a matching number of physical cells.
Requires cell topology with a reference cell. See Section Reference-to-physical cell mapping for definition of the mapping function. Presently supported cell topologies are
- 1D:
Line<2> - 2D:
Triangle<3>,Triangle<6>,Quadrilateral<4>,Quadrilateral<9> - 3D:
Tetrahedron<4>,Tetrahedron<10>,Hexahedron<8>,Hexahedron<27>
- Warning
- The array
refPointsrepresents an arbitrary set of points in the reference frame that are not required to be in the reference cell corresponding to the specified cell topology. As a result, the images of these points under a given reference-to-physical map are not necessarily contained in the physical cell that is the image of the reference cell under that map. CellTools provides several inclusion tests methods to check whether or not the points are inside a reference cell.
- Parameters
-
physPoints [out] - rank-3 array with dimensions (C,P,D) with the images of the ref. points refPoints [in] - rank-3/2 array with dimensions (C,P,D)/(P,D) with points in reference frame cellWorkset [in] - rank-3 array with dimensions (C,N,D) with the nodes of the cell workset cellTopo [in] - cell topology of the cells stored in cellWorkset
Definition at line 1051 of file Intrepid2_CellTools.hpp.
References Intrepid2::CellTools< DeviceType >::mapToPhysicalFrame().
|
static |
Computes 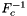 , the inverse of the reference-to-physical frame map using a default initial guess.
, the inverse of the reference-to-physical frame map using a default initial guess.
Applies 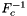 for all cells in a cell workset to multiple point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array. Returns a rank-3 (C,P,D) array such that
for all cells in a cell workset to multiple point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array. Returns a rank-3 (C,P,D) array such that
![\[ \mbox{refPoints}(c,p,d) = \Big(F^{-1}_c(physPoint(c,p,*)) \Big)_d \]](form_76.png)
Requires cell topology with a reference cell. See Section Reference-to-physical cell mapping for definition of the mapping function. Presently supported cell topologies are
- 1D:
Line<2> - 2D:
Triangle<3>,Triangle<6>,Quadrilateral<4>,Quadrilateral<9> - 3D:
Tetrahedron<4>,Tetrahedron<10>,Hexahedron<8>,Hexahedron<27>
- Warning
- Computation of the inverse map in this method uses default selection of the initial guess based on cell topology:
Linetopologies: line center (0)Triangletopologies: the point (1/3, 1/3)Quadrilateraltopologies: the point (0, 0)Tetrahedrontopologies: the point (1/6, 1/6, 1/6)Hexahedrontopologies: the point (0, 0, 0)Wedgetopologies: the point (1/2, 1/2, 0) For some cells with extended topologies, these initial guesses may not be good enough for Newton's method to converge in the allotted number of iterations. A version of this method with user-supplied initial guesses is also available.
-
The array
physPointsrepresents an arbitrary set (or sets) of points in the physical frame that are not required to belong in the physical cell (cells) that define(s) the reference to physical mapping. As a result, the images of these points in the reference frame are not necessarily contained in the reference cell corresponding to the specified cell topology.
- Parameters
-
refPoints [out] - rank-3 array with dimensions (C,P,D) with the images of the physical points physPoints [in] - rank-3 array with dimensions (C,P,D) with points in physical frame worksetCell [in] - rank-3 array with dimensions (C,N,D) with the nodes of the cell workset cellTopo [in] - cell topology of the cells stored in cellWorkset
Definition at line 39 of file Intrepid2_CellToolsDefPhysToRef.hpp.
Referenced by Intrepid2::PointTools::getWarpBlendLatticeTetrahedron(), and Intrepid2::PointTools::getWarpBlendLatticeTriangle().
|
static |
Computation of 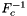 , the inverse of the reference-to-physical frame map using user-supplied initial guess.
, the inverse of the reference-to-physical frame map using user-supplied initial guess.
Applies 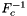 for all cells in a cell workset to multiple point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array. Returns a rank-3 (C,P,D) array such that
for all cells in a cell workset to multiple point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array. Returns a rank-3 (C,P,D) array such that
![\[ \mbox{refPoints}(c,p,d) = \Big(F^{-1}_c(physPoint(c,p,*)) \Big)_d \]](form_76.png)
Requires pointer to HGrad basis that defines reference to physical cell mapping. See Section Reference-to-physical cell mapping for definition of the mapping function.
- Warning
- The array
physPointsrepresents an arbitrary set (or sets) of points in the physical frame that are not required to belong in the physical cell (cells) that define(s) the reference to physical mapping. As a result, the images of these points in the reference frame are not necessarily contained in the reference cell corresponding to the specified cell topology.
- Parameters
-
refPoints [out] - rank-3/2 array with dimensions (C,P,D)/(P,D) with the images of the physical points initGuess [in] - rank-3/2 array with dimensions (C,P,D)/(P,D) with the initial guesses for each point physPoints [in] - rank-3/2 array with dimensions (C,P,D)/(P,D) with points in physical frame worksetCell [in] - rank-3 array with dimensions (C,N,D) with the nodes of the cell workset basis [in] - pointer to HGrad basis used for reference to physical cell mapping
Definition at line 90 of file Intrepid2_CellToolsDefPhysToRef.hpp.
References Intrepid2::Parameters::MaxNewton.
Referenced by Intrepid2::CellTools< DeviceType >::mapToReferenceFrameInitGuess().
|
inlinestatic |
Computation of 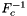 , the inverse of the reference-to-physical frame map using user-supplied initial guess.
, the inverse of the reference-to-physical frame map using user-supplied initial guess.
Applies 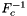 for all cells in a cell workset to multiple point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array. Returns a rank-3 (C,P,D) array such that
for all cells in a cell workset to multiple point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array. Returns a rank-3 (C,P,D) array such that
![\[ \mbox{refPoints}(c,p,d) = \Big(F^{-1}_c(physPoint(c,p,*)) \Big)_d \]](form_76.png)
Requires cell topology with a reference cell. See Section Reference-to-physical cell mapping for definition of the mapping function. Presently supported cell topologies are
- 1D:
Line<2> - 2D:
Triangle<3>,Triangle<6>,Quadrilateral<4>,Quadrilateral<9> - 3D:
Tetrahedron<4>,Tetrahedron<10>,Hexahedron<8>,Hexahedron<27>
- Warning
- The array
physPointsrepresents an arbitrary set (or sets) of points in the physical frame that are not required to belong in the physical cell (cells) that define(s) the reference to physical mapping. As a result, the images of these points in the reference frame are not necessarily contained in the reference cell corresponding to the specified cell topology.
- Parameters
-
refPoints [out] - rank-3/2 array with dimensions (C,P,D)/(P,D) with the images of the physical points initGuess [in] - rank-3/2 array with dimensions (C,P,D)/(P,D) with the initial guesses for each point physPoints [in] - rank-3/2 array with dimensions (C,P,D)/(P,D) with points in physical frame worksetCell [in] - rank-3 array with dimensions (C,N,D) with the nodes of the cell workset cellTopo [in] - cell topology of the cells stored in cellWorkset
Definition at line 1283 of file Intrepid2_CellTools.hpp.
References Intrepid2::CellTools< DeviceType >::mapToReferenceFrameInitGuess().
|
static |
Computes parameterization maps of 1- and 2-subcells of reference cells.
Applies 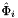 , the parametrization map of a subcell
, the parametrization map of a subcell 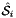 from a given reference cell, to a set of points in the parametrization domain R of
from a given reference cell, to a set of points in the parametrization domain R of 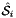 . Returns a rank-2 array with dimensions (P,D) where for 1-subcells:
. Returns a rank-2 array with dimensions (P,D) where for 1-subcells:
![\[ {subcellPoints}(p,*) = \hat{\Phi}_i(t_p) \quad\mbox{and}\quad \hat{\Phi}_i(t_p) = \left\{ \begin{array}{ll} (\hat{x}(t_p),\hat{y}(t_p),\hat{z}(t_p)) & \mbox{for 3D parent cells}\\[1.5ex] (\hat{x}(t_p),\hat{y}(t_p)) & \mbox{for 2D parent cells} \end{array} \right. \quad t_p \in R = [-1,1] \,; \]](form_71.png)
for 2-subcells:
![\[ {subcellPoints}(p,*) = \hat{\Phi}_i(u_p,v_p)\quad\mbox{and}\quad \hat{\Phi}_i(u_p,v_p) = (\hat{x}(u_p,v_p), \hat{y}(u_p,v_p), \hat{z}(u_p, v_p)) \quad (u_p,v_p)\in R \]](form_72.png)
and
![\[ R = \left\{\begin{array}{rl} \{(0,0),(1,0),(0,1)\} & \mbox{if face is Triangle} \\[1ex] [-1,1]\times [-1,1] & \mbox{if face is Quadrilateral} \end{array}\right. \]](form_73.png)
- Remarks
- Parametrization of 1-subcells is defined for all 2D and 3D cell topologies with reference cells, including special 2D and 3D topologies such as shell and beams.
- Parametrization of 2-subcells is defined for all 3D cell topologies with reference cells, including special 3D topologies such as shells.
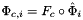 of each subcell in the workset to
of each subcell in the workset to paramPoints. Here c is ordinal of a parent cell, relative to subcell workset, and i is subcell ordinal, relative to a reference cell, of the subcell workset. See Section Subcell worksets for definition of subcell workset and Section Parametrization of physical 1- and 2-subcells for definition of parametrization maps.
- Parameters
-
refSubcellPoints [out] - rank-2 (P,D1) array with images of parameter space points paramPoints [in] - rank-2 (P,D2) array with points in 1D or 2D parameter domain subcellDim [in] - dimension of the subcell where points are mapped to subcellOrd [in] - subcell ordinal parentCell [in] - cell topology of the parent cell.
Referenced by Intrepid2::Basis_HCURL_TET_In_FEM< DeviceType, outputValueType, pointValueType >::Basis_HCURL_TET_In_FEM(), Intrepid2::Basis_HCURL_TRI_In_FEM< DeviceType, outputValueType, pointValueType >::Basis_HCURL_TRI_In_FEM(), Intrepid2::Basis_HDIV_TET_In_FEM< DeviceType, outputValueType, pointValueType >::Basis_HDIV_TET_In_FEM(), Intrepid2::Basis_HDIV_TRI_In_FEM< DeviceType, outputValueType, pointValueType >::Basis_HDIV_TRI_In_FEM(), Intrepid2::Basis_HGRAD_TET_Cn_FEM< DeviceType, outputValueType, pointValueType >::Basis_HGRAD_TET_Cn_FEM(), Intrepid2::ProjectionStruct< DeviceType, ValueType >::createHCurlProjectionStruct(), Intrepid2::ProjectionStruct< DeviceType, ValueType >::createHDivProjectionStruct(), Intrepid2::ProjectionStruct< DeviceType, ValueType >::createHGradProjectionStruct(), Intrepid2::ProjectionStruct< DeviceType, ValueType >::createL2ProjectionStruct(), Intrepid2::CubatureControlVolumeBoundary< DeviceType, pointValueType, weightValueType >::CubatureControlVolumeBoundary(), and Intrepid2::CubatureControlVolumeSide< DeviceType, pointValueType, weightValueType >::CubatureControlVolumeSide().
|
static |
Computes parameterization maps of 1- and 2-subcells of reference cells.
Overload of the previous function (see explanation above) where the subcell parametrization is used instead of passing the parent cell topology.
|
static |
Computes parameterization maps of 1- and 2-subcells of reference cells.
Overload of the previous function (see explanation above) where the subcellOrd is a rank-1 array (P).
|
static |
Computes the Jacobian matrix DF of the reference-to-physical frame map F.
There are two use cases:
- Computes Jacobians
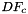 of the reference-to-physical map for all physical cells in a cell workset on a single set of reference points stored in a rank-2 (P,D) array;
of the reference-to-physical map for all physical cells in a cell workset on a single set of reference points stored in a rank-2 (P,D) array; - Computes Jacobians
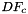 of the reference-to-physical map for all physical cells in a cell workset on multiple reference point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array;
of the reference-to-physical map for all physical cells in a cell workset on multiple reference point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array;
For a single point set in a rank-2 array (P,D) returns a rank-4 (C,P,D,D) array such that
![\[ \mbox{jacobian}(c,p,i,j) = [DF_{c}(\mbox{points}(p))]_{ij} \quad c=0,\ldots, C \]](form_1.png)
For multiple sets of reference points in a rank-3 (C,P,D) array returns rank-4 (C,P,D,D) array such that
![\[ \mbox{jacobian}(c,p,i,j) = [DF_{c}(\mbox{points}(c,p))]_{ij} \quad c=0,\ldots, C \]](form_2.png)
Requires pointer to HGrad basis that defines reference to physical cell mapping. See Section \ref sec_cell_topology_ref_map_DF for definition of the Jacobian. \warning The points are not required to be in the reference cell associated with the specified cell topology. CellTools provides several inclusion tests methods to check whether or not the points are inside a reference cell.
- Parameters
-
jacobian [out] - rank-4 array with dimensions (C,P,D,D) with the Jacobians points [in] - rank-2/3 array with dimensions (P,D)/(C,P,D) with the evaluation points cellWorkset [in] - rank-3 container with logical dimensions (C,N,D) with the nodes of the cell workset basis [in] - HGrad basis for reference to physical cell mapping startCell [in] - first cell index in cellWorkset for which we should compute the Jacobian; corresponds to the 0 index in Jacobian and/or points container. Default: 0. endCell [in] - first cell index in cellWorkset that we do not process; endCell - startCell must equal the extent of the Jacobian container in dimension 0. Default: -1, a special value that indicates the extent of the cellWorkset should be used.
Definition at line 799 of file Intrepid2_CellToolsDefJacobian.hpp.
Referenced by Intrepid2::CubatureControlVolumeBoundary< DeviceType, pointValueType, weightValueType >::getCubature(), Intrepid2::CubatureControlVolume< DeviceType, pointValueType, weightValueType >::getCubature(), Intrepid2::CubatureControlVolumeSide< DeviceType, pointValueType, weightValueType >::getCubature(), Intrepid2::CellTools< DeviceType >::setJacobian(), and Intrepid2::CellGeometry< PointScalar, spaceDim, DeviceType >::setJacobianDataPrivate().
|
static |
Computes the Jacobian matrix DF of the reference-to-physical frame map F.
There are two use cases:
- Computes Jacobians
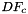 of the reference-to-physical map for all physical cells in a cell workset on a single set of reference points stored in a rank-2 (P,D) array;
of the reference-to-physical map for all physical cells in a cell workset on a single set of reference points stored in a rank-2 (P,D) array; - Computes Jacobians
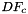 of the reference-to-physical map for all physical cells in a cell workset on multiple reference point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array;
of the reference-to-physical map for all physical cells in a cell workset on multiple reference point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array;
For a single point set in a rank-2 array (P,D) returns a rank-4 (C,P,D,D) array such that
![\[ \mbox{jacobian}(c,p,i,j) = [DF_{c}(\mbox{points}(p))]_{ij} \quad c=0,\ldots, C \]](form_1.png)
For multiple sets of reference points in a rank-3 (C,P,D) array returns rank-4 (C,P,D,D) array such that
![\[ \mbox{jacobian}(c,p,i,j) = [DF_{c}(\mbox{points}(c,p))]_{ij} \quad c=0,\ldots, C \]](form_2.png)
Requires pointer to HGrad basis that defines reference to physical cell mapping. See Section \ref sec_cell_topology_ref_map_DF for definition of the Jacobian. \warning The points are not required to be in the reference cell associated with the specified cell topology. CellTools provides several inclusion tests methods to check whether or not the points are inside a reference cell.
- Parameters
-
jacobian [out] - rank-4 array with dimensions (C,P,D,D) with the Jacobians cellWorkset [in] - rank-3 container with logical dimensions (C,N,D) with the nodes of the cell workset gradients [in] - rank-3/4 array with dimensions (N,P,D)/(C,N,P,D) with the gradients of the physical-to-cell mapping startCell [in] - first cell index in cellWorkset for which we should compute the Jacobian; corresponds to the 0 index in Jacobian and/or points container. Default: 0. endCell [in] - first cell index in cellWorkset that we do not process; endCell - startCell must equal the extent of the Jacobian container in dimension 0. Default: -1, a special value that indicates the extent of the cellWorkset should be used.
Definition at line 771 of file Intrepid2_CellToolsDefJacobian.hpp.
|
inlinestatic |
Computes the Jacobian matrix DF of the reference-to-physical frame map F.
There are two use cases:
- Computes Jacobians
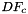 of the reference-to-physical map for all physical cells in a cell workset on a single set of reference points stored in a rank-2 (P,D) array;
of the reference-to-physical map for all physical cells in a cell workset on a single set of reference points stored in a rank-2 (P,D) array; - Computes Jacobians
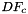 of the reference-to-physical map for all physical cells in a cell workset on multiple reference point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array;
of the reference-to-physical map for all physical cells in a cell workset on multiple reference point sets having the same number of points, indexed by cell ordinal, and stored in a rank-3 (C,P,D) array;
For a single point set in a rank-2 array (P,D) returns a rank-4 (C,P,D,D) array such that
![\[ \mbox{jacobian}(c,p,i,j) = [DF_{c}(\mbox{points}(p))]_{ij} \quad c=0,\ldots, C \]](form_1.png)
For multiple sets of reference points in a rank-3 (C,P,D) array returns rank-4 (C,P,D,D) array such that
![\[ \mbox{jacobian}(c,p,i,j) = [DF_{c}(\mbox{points}(c,p))]_{ij} \quad c=0,\ldots, C \]](form_2.png)
Requires cell topology with a reference cell. See Section \ref sec_cell_topology_ref_map_DF for definition of the Jacobian. \warning The points are not required to be in the reference cell associated with the specified cell topology. CellTools provides several inclusion tests methods to check whether or not the points are inside a reference cell.
- Parameters
-
jacobian [out] - rank-4 array with dimensions (C,P,D,D) with the Jacobians points [in] - rank-2/3 array with dimensions (P,D)/(C,P,D) with the evaluation points cellWorkset [in] - rank-3 array with dimensions (C,N,D) with the nodes of the cell workset cellTopo [in] - cell topology of the cells stored in cellWorkset
Definition at line 284 of file Intrepid2_CellTools.hpp.
References Intrepid2::CellTools< DeviceType >::setJacobian().
|
static |
Computes the determinant of the Jacobian matrix DF of the reference-to-physical frame map F.
Returns rank-2 array with dimensions (C,P) such that
![\[ \mbox{jacobianDet}(c,p) = \mbox{det}(\mbox{jacobian}(c,p,*,*)) \quad c=0,\ldots, C \]](form_4.png)
- Parameters
-
jacobianDet [out] - rank-2 array with dimensions (C,P) with Jacobian determinants jacobian [in] - rank-4 array with dimensions (C,P,D,D) with the Jacobians
Definition at line 874 of file Intrepid2_CellToolsDefJacobian.hpp.
References Intrepid2::RealSpaceTools< DeviceType >::det().
Referenced by Intrepid2::CubatureControlVolumeBoundary< DeviceType, pointValueType, weightValueType >::getCubature(), and Intrepid2::CubatureControlVolume< DeviceType, pointValueType, weightValueType >::getCubature().
|
static |
Computes determinants corresponding to the Jacobians in the Data container provided.
- Parameters
-
jacobianDet [out] - data with shape (C,P), as returned by CellTools::allocateJacobianDet() jacobian [in] - data with shape (C,P,D,D), as returned by CellGeometry::allocateJacobianData()
Definition at line 165 of file Intrepid2_CellToolsDefJacobian.hpp.
References Intrepid2::Data< DataScalar, DeviceType >::blockPlusDiagonalLastNonDiagonal(), Intrepid2::RealSpaceTools< DeviceType >::det(), Intrepid2::Data< DataScalar, DeviceType >::extent_int(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView1(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView2(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView3(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView4(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingViewRank(), Intrepid2::Data< DataScalar, DeviceType >::getVariationTypes(), and INTREPID2_TEST_FOR_EXCEPTION_DEVICE_SAFE.
|
static |
Computes reciprocals of determinants corresponding to the Jacobians in the Data container provided.
- Parameters
-
jacobianDetInv [out] - data with shape (C,P), as returned by CellTools::allocateJacobianDet() jacobian [in] - data with shape (C,P,D,D), as returned by CellGeometry::allocateJacobianData()
Definition at line 432 of file Intrepid2_CellToolsDefJacobian.hpp.
References Intrepid2::Data< DataScalar, DeviceType >::allocateConstantData(), and Intrepid2::Data< DataScalar, DeviceType >::storeInPlaceQuotient().
|
static |
Multiplies the Jacobian with shape (C,P,D,D) by the reciprocals of the determinants, with shape (C,P), entrywise.
- Parameters
-
jacobianDividedByDet [out] - data container with shape (C,P,D,D), as returned by CellTools::allocateJacobianInv() jacobian [in] - data container with shape (C,P,D,D), as returned by CellTools::allocateJacobianInv() jacobianDetInv [in] - data with shape (C,P,D,D), as returned by CellGeometry::allocateJacobianData()
|
static |
Computes the inverse of the Jacobian matrix DF of the reference-to-physical frame map F.
Returns rank-4 array with dimensions (C,P,D,D) such that
![\[ \mbox{jacobianInv}(c,p,*,*) = \mbox{jacobian}^{-1}(c,p,*,*) \quad c = 0,\ldots, C \]](form_3.png)
- Parameters
-
jacobianInv [out] - rank-4 array with dimensions (C,P,D,D) with the inverse Jacobians jacobian [in] - rank-4 array with dimensions (C,P,D,D) with the Jacobians
Definition at line 861 of file Intrepid2_CellToolsDefJacobian.hpp.
References Intrepid2::RealSpaceTools< DeviceType >::inverse().
|
static |
Computes determinants corresponding to the Jacobians in the Data container provided.
- Parameters
-
jacobianInv [out] - data container with shape (C,P,D,D), as returned by CellTools::allocateJacobianInv() jacobian [in] - data with shape (C,P,D,D), as returned by CellGeometry::allocateJacobianData()
Definition at line 442 of file Intrepid2_CellToolsDefJacobian.hpp.
References Intrepid2::Data< DataScalar, DeviceType >::blockPlusDiagonalLastNonDiagonal(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView1(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView2(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView3(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingView4(), Intrepid2::Data< DataScalar, DeviceType >::getUnderlyingViewRank(), Intrepid2::Data< DataScalar, DeviceType >::getVariationTypes(), INTREPID2_TEST_FOR_EXCEPTION_DEVICE_SAFE, and Intrepid2::RealSpaceTools< DeviceType >::inverse().
The documentation for this class was generated from the following files:
- http://docs.trilinos.org/dev/packages/intrepid2/src/Cell/Intrepid2_CellTools.hpp
- http://docs.trilinos.org/dev/packages/intrepid2/src/Cell/Intrepid2_CellToolsDefControlVolume.hpp
- http://docs.trilinos.org/dev/packages/intrepid2/src/Cell/Intrepid2_CellToolsDefInclusion.hpp
- http://docs.trilinos.org/dev/packages/intrepid2/src/Cell/Intrepid2_CellToolsDefJacobian.hpp
- http://docs.trilinos.org/dev/packages/intrepid2/src/Cell/Intrepid2_CellToolsDefNodeInfo.hpp
- http://docs.trilinos.org/dev/packages/intrepid2/src/Cell/Intrepid2_CellToolsDefPhysToRef.hpp
- http://docs.trilinos.org/dev/packages/intrepid2/src/Cell/Intrepid2_CellToolsDefRefToPhys.hpp
 1.8.5
1.8.5