
|
Many third-party solvers support a vast amount of parameters to control factorization and solution. An effort has been made in Amesos2 to expose to users as many of those parameters as reasonably possible. Not all parameters may be supported, but if there is one that you would like to have exposed, then contact the Amesos2 developers and we may be able to work something out for you.
amesos2_parameters
The following parameters are currently acted upon by Amesos2 solvers:
-
"Transpose": {true|false}. Iftrue, tells the solver to solve for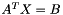
We plan in the future to support the following parameters:
-
"Reindex": {true|false}. Put the matrix row and column indices into the range [0..n]. -
"AddZeroToDiag": {true|false}. -
"AddToDiag":string. Where the given string is a representation of a scalar value to add to all diagonal entries of the matrix before factorization.
Solver-specific Parameters
SuperLU
Currently, the following SuperLU parameters/options are recognized and acted upon:
-
"Trans": {"NOTRANS"|"TRANS"|"CONJ"}. Specifies whether to solve with the transpose system. -
"Equil": {true|false}. Specifies whether the solver to equilibrate the matrix before solving. -
"IterRefine": {"NO"|"SLU_SINGLE"|"SLU_DOUBLE"|"EXTRA"}. Specifies whether to perform iterative refinement, and in what precision to compute the residual. -
"SymmetricMode": {true|false}. -
"DiagPivotThresh":doublevalue. Specifies the threshold used for a diagonal to be considered an acceptable pivot. -
"ColPerm"which takes one of the following:-
"NATURAL": natural ordering. -
"MMD_AT_PLUS_A": minimum degree ordering on the structure of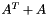 .
. -
"MMD_ATA": minimum degree ordering on the structure of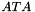 .
. -
"COLAMD": approximate minimum degree column ordering. (default)
-
-
"ILU_Flag": {false|true}. If true, run ILU routines. -
"ILU_DropTol":doublevalue. ILUT drop tolerance. -
"ILU_FillFactor":doublevalue. ILUT fill factor. -
"ILU_Norm": {"INF_NORM"|"ONE_NORM"|"TWO_NORM"}. -
"ILU_MILU": {"SILU"|"SMILU_1"|"SMILU_2"|"SMILU_3"}. Type of modified ILU algorithm to use. -
"ILU_FillTol":doublevalue. ILUT fill tolerance.
SuperLU_MT
The following SuperLU_MT parameters/options are recognized:
-
"nprocs"(int) : Specifies the number of threads to be spawned. Default is 1. -
"trans": {"NOTRANS"|"TRANS"|"CONJ"}. Will also recognize the"Transpose": {true|false} option which is equivalent to"TRANS"and"NOTRANS", respectively. -
"panel_size"(int) : Specifies the number of consecutive columns to be treated as a unit of task. -
"relax"(int) : Specifies the number of columns to be grouped as a relaxed supernode. -
"Equil": {true|false}. Specifies whether the solver to equilibrate the matrix before solving. -
"SymmetricMode": {true|false}. Specifies whether to use the symmetric mode. -
"DiagPivotThresh":doublevalue. Specifies the threshold used for a diagonal to be considered an acceptable pivot. -
"ColPerm"which takes one of the following:-
"NATURAL": natural ordering. -
"MMD_AT_PLUS_A": minimum degree ordering on the structure of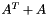 .
. -
"MMD_ATA": minimum degree ordering on the structure of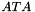 .
. -
"COLAMD": approximate minimum degree column ordering. (default)
-
Note that the nprocs, panel_size, and relax options are recognized by SuperLU_MT but not by SuperLU. Note also that it is no typo in "trans", it really is lower-case (as opposed to upper-case in SuperLU)
SuperLU_DIST
Currently, the following SuperLU_DIST parameters/options are recognized:
-
"npcol"(int) and "nprow"(int) : Specified together, these parameters set the size of the SuperLU_DIST processor grid tonprowrows bynpcolcolumns. If these parameters are not set, the SuperLU_DIST interface uses a heuristic to pick the grid dimensions based on the number of processors in the matrix' communicator. -
"ColPerm"which takes one of the following:-
"NATURAL": natural column ordering. -
"PARMETIS": use the ParMETIS TPL to order the columns. (default)
-
-
"ReplaceTinyPivot": {true|false}. Specifies whether to replace tiny diagonals with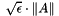 during LU factorization. (default:
during LU factorization. (default: true)
Pardiso MKL
The Pardiso MKL parameters that are currently recognized are:
-
"IPARM(2)" -
"IPARM(4)" -
"IPARM(8)" -
"IPARM(10)" -
"IPARM(12)" -
"IPARM(18)" -
"IPARM(24)" -
"IPARM(25)" -
"IPARM(60)"
Please see the Pardiso MKL documentation for a summary of the meaning and valid values for each parameter.
LAPACK
The LAPACK interface supports the following parameters:
-
"Equilibrate"(bool) : equilibrate the matrix before factorization. The default istrue.
 1.8.5
1.8.5